Ribbon diagram of the restriction enzyme EcoRI
BIOL 1406
PreLab 9a.3
How do restriction enzymes cut DNA into fragments and how can these fragments be analyzed using agarose gel electrophoresis?
|
|
|
Ribbon diagram of the restriction enzyme EcoRI |
Restriction enzymes are naturally
occurring enzymes that cut DNA. Many restriction enzymes have been isolated
from bacteria, providing a valuable tool for molecular biologists. Each
restriction enzyme cuts DNA only where a specific sequence of base pairs
occurs. For example, these are the sequences cut by the restriction enzymes
BamHI and XmaI.: 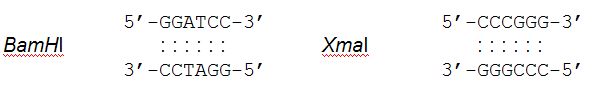 An important difference among restriction enzymes is whether the cuts they make produce “blunt ends” or “sticky ends.” For example, HaeIII cuts this sequence:  Because these 2 fragments do not have any unpaired bases, we say they have “blunt ends.” EcoRI, on the other hand, cuts this sequence: 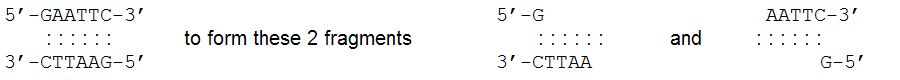 Because these 2 fragments have unpaired bases which can potentially pair with complementary bases, we say they have “sticky ends.” |
If we cut DNA from two different sources with the same restriction enzyme, DNA
fragments with complementary sticky ends are formed:
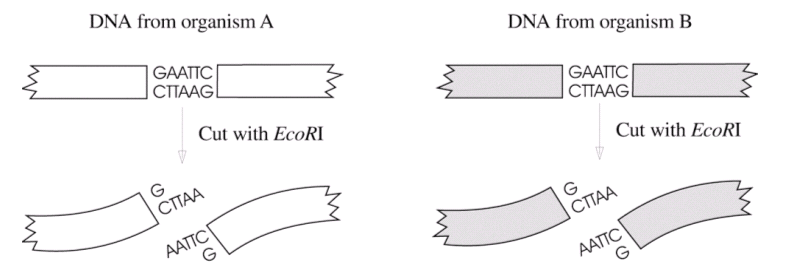 |
If the DNA fragments are mixed together, they will join together at random,
provided their unpaired bases match. Therefore, sometimes pieces of DNA from the
same source will join back together, and sometimes pieces of DNA from different
sources will join together, as shown below.
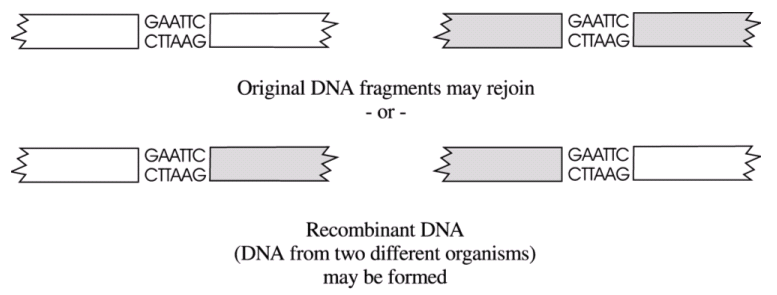 |
Hydrogen-bonding between complementary bases is not sufficient to completely
rejoin the DNA fragments. The broken bonds between the deoxyribose and phosphate
groups that form the “side-rails” of the DNA double helix (the phosphodiester
linkages) must also be repaired. DNA ligase is the enzyme, also isolated from
bacteria, that catalyzes this reaction.
In this lab exercise, you will be given 3 unknown samples labeled A, B, and C. One sample has no plasmids at all, one sample has normal pUC18 plasmids, and one sample has recombinant pUC18 plasmids. Your job is to determine which plasmids, if any, are present in each sample. To do this, you will carry out 2 experiments:
|
|
|
| Your Turn | |
| What is a recombinant plasmid? | Check your answer. |
| What are restriction enzymes? | Check your answer. |
| What is DNA ligase? | Check your answer. |
| If a pUC18 plasmid is cut in only one location by the restriction enzyme EcoRI, it will be changed from a circular form to a form. | Check your answer. |
| Imagine that both pUC18 plasmids and phage DNA
are cut with EcoRI. Each plasmid is cut in one location to form
a linear fragment and
each phage is cut in one location to form a linear fragment. Then all
of the linear fragments are mixed together. In the spaces below, list the five possible ways circular pieces of DNA can
form by joining the ends of either one or two of these linear DNA fragments. Note
that the 2 ends of one fragment can join to form a circle, or two different
fragments can join to make a circle: 1. 2. 3. 4. 5. |
Check your answer. |
| Which of the 5 possible combinations
represent a recombinant plasmid?
|
Check your answer. |
| Which of the 5 possible combinations
represent plasmids that are not recombinant?
|
Check your answer. |
| Which of the 5 possible combinations
represent circular pieces of DNA that do not include a plasmid?
|
Check your answer. |
| After cutting recombinant and
non-recombinant plasmids into fragments with the restriction enzyme EcoRI, which procedure will you use
to separate the fragments and determine their size?
|
Check your answer. |
Close this browser window to return
to Blackboard and complete the practice quiz and assessment quiz.