- One unknown solution contains no DNA at all.
- One unknown solution contains normal pUC18 plasmids. These plasmids contain a
gene for ampicillin resistance, and the lacZ gene. Recall that the lacZ gene
codes for the enzyme β–galactosidase. This enzyme is normally used by
E. coli
cells to digest lactose. However, when bacteria with this gene are grown in a
medium containing a synthetic analog of lactose called Xgal, the enzyme digests
Xgal and produces a blue compound.
- One unknown solution contains recombinant pUC18 plasmids. Like the normal pUC18 plasmids, the recombinant pUC18 plasmids have the lacZ gene and the gene for ampicillin resistance. However, the recombinant pUC18 plasmids also contain a piece of foreign DNA from a phage named λ. In order to insert the phage DNA into pUC18, pUC18 was cut with a restriction enzyme called EcoRI. EcoRI cuts pUC18 at a site located within the lacZ gene. Therefore, the extra piece of phage DNA was inserted into the lacZ gene and inactivated it. This means no ß–galactosidase will be produced. Therefore, Xgal is not digested, no blue color is produced, and any colonies that grow will remain white.
After incubating competent E. coli cells with the 3 unknown solutions described above, the cells will be inoculated on 3 separate Petri dishes containing nutrient agar with ampicillin and Xgal. The Petri dishes will be incubated for several days at 37° C to allow the bacteria to replicate. When placed on nutrient agar, one bacterial cell can divide over and over until there are millions of descendants on the plate. Although one bacterium is not visible to the naked eye, the millions of descendants piled together appear as a shiny dot, called a bacterial colony, on the surface of the agar.
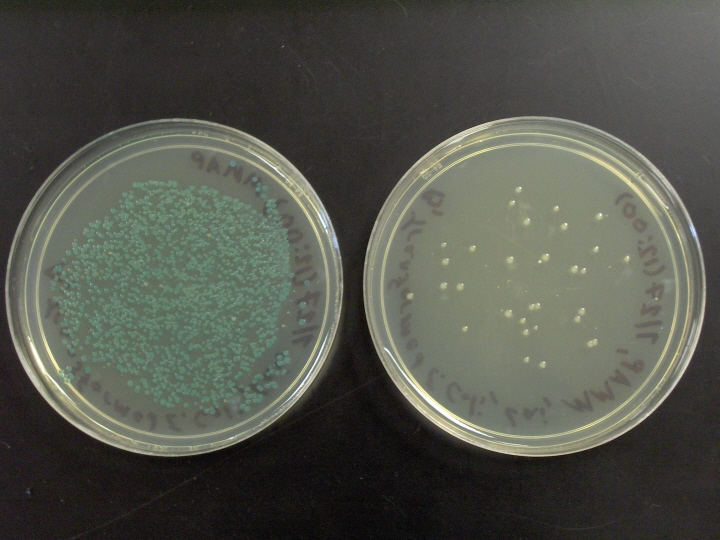
Blue and white colonies
of E. coli growing on nutrient
agar that contains X-gal