BIOL 1406
PreLab
9b.1
After cutting DNA with a restriction enzyme, how can I use
agarose gel electrophoresis to analyze the DNA fragments?
During exercise 9a, you were given 3 unknown plasmid samples labeled A, B, and
C. These samples were used to carry out 2 experiments:
- Competent E. coli cells were mixed with the 3 unknown samples in order to
allow transformation to take place. The E. coli cells were then plated on
nutrient agar containing ampicillin and Xgal.
- The restriction enzyme EcoRI was mixed with the 3 unknown samples in order to
cut any DNA present into fragments. The resulting DNA fragments were then
separated using agarose gel electrophoresis.
In this lab, you will view the results of these 2 experiments in order
to determine which unknown sample had no plasmids, which sample had
non-recombinant pUC18
plasmids, and which sample had recombinant pUC18 plasmids. In addition, you will
use the results of agarose gel electrophoresis to estimate the size of the
fragments that were produced when EcoRI digested the DNA in the 3 unknown
samples.
 |
The separation of DNA molecules by agarose gel electrophoresis is similar to the
separation of proteins by SDS-PAGE electrophoresis. In both cases, the molecules
to be separated are loaded onto a gel and then subjected to an electrical field.
The cathode (negative electrode) attracts positively charged molecules, while
the anode (positive electrode) attracts negatively charged molecules. This
attraction causes the molecules in the mixture to migrate through the gel at different rates depending on their size, shape, chemical composition,
and electrical charge. As the molecules migrate at different rates, they
gradually separate from each other.
The main difference between protein electrophoresis and DNA electrophoresis lies
in the support medium. While polyacrylamide gels are generally used for protein
separations, agarose gels are commonly used for DNA separations. Agarose is a
polysaccharide that is extracted from seaweed. Agarose gels are cast by
dissolving agarose in a Tris-buffered solvent at a high temperature, and then
pouring the solution into a horizontal tray and allowing it to cool. A comb is
inserted into the cooling gel so that wells are formed as the gel solidifies.
The cooled gel is then transferred into a gel box where it is submerged in a
Tris-buffered electrode solution before loading DNA samples into the wells.
Since these gels are run horizontally and beneath the electrode buffer, they are
sometimes referred to as “horizontal gels” or “submarine gels”. |
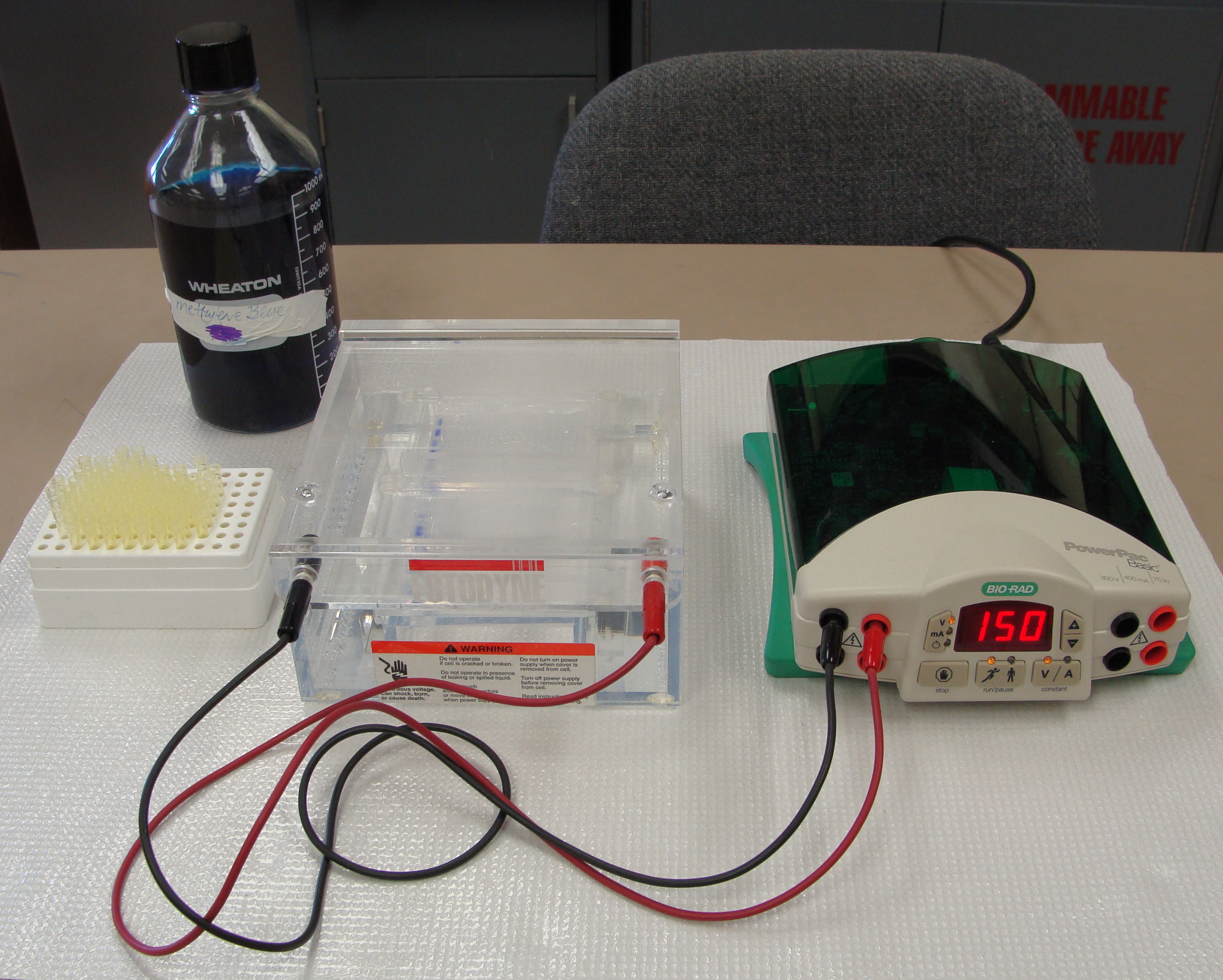
|
Lab set-up for agarose gel
electrophoresis |
DNA molecules will migrate towards the anode during electrophoresis because they
have a high concentration of negatively-charged phosphate groups in the
sugar-phosphate backbone of the double helix. These negative charges provide a
fairly uniform charge-to-mass ratio among all DNA molecules. Because of this
uniform ratio, the migration rate of DNA molecules during agarose gel
electrophoresis is almost entirely a function of size. As with protein
electrophoresis, smaller molecules move through the gel more rapidly than larger
molecules. Scientists generally measure the size of DNA molecules in terms of
number of base pairs (bp) rather than molecular weight (MW).
DNA, like most proteins, is colorless. Therefore, before DNA samples are loaded
onto an agarose gel, they are mixed with a sample buffer that includes a blue
tracking dye. During electrophoresis, the tracking dye migrates very rapidly
through the gel, along with the smallest of DNA fragments. When the dye
approaches the end of the gel, you know it is time to stop the electrophoresis.
Glycerol is also included in the sample buffer in order to make the sample
denser so that it settles into the well as it is loaded.
After electrophoresis is complete, the gel is stained so that the DNA bands can
be seen. The most commonly used dye for staining DNA is ethidium bromide. DNA
bound to ethidium bromide fluoresces strongly under ultraviolet light, so the
gels are viewed and photographed using ultraviolet light. However, because both
ethidium bromide and ultraviolet light are mutagens, you will be using a less
sensitive but safer stain for your DNA gels: methylene blue. Methylene blue is a
blue-colored stain that binds to DNA in the same way that Coomassie Blue binds
to proteins. The blue color is easily seen using normal visible light
Recall that during SDS-PAGE electrophoresis there is a linear relationship
between the migration distance of the proteins and log of MW. Similarly, during
agarose gel electrophoresis there is a linear relationship between the migration
distance of the DNA molecules and log of bp (number of base pairs.) Because of
this, agarose gel electrophoresis can be used to estimate the size (number of
base pairs) of DNA molecules. In order to do this, a standard curve that shows
the relationship between migration distance through the gel and log of bp must
be generated. The standard curve is generated by measuring the migration
distance of several DNA molecules of known size (number of base pairs), plotting
a scatter diagram of migration distance vs. log of bp, and then using linear
regression to determine the equation of the “best fit” straight line for the
data points. Once this is done, you can substitute the migration distance of any
DNA molecule on your gel into the linear regression equation and then calculate
the log of bp for that molecule. |
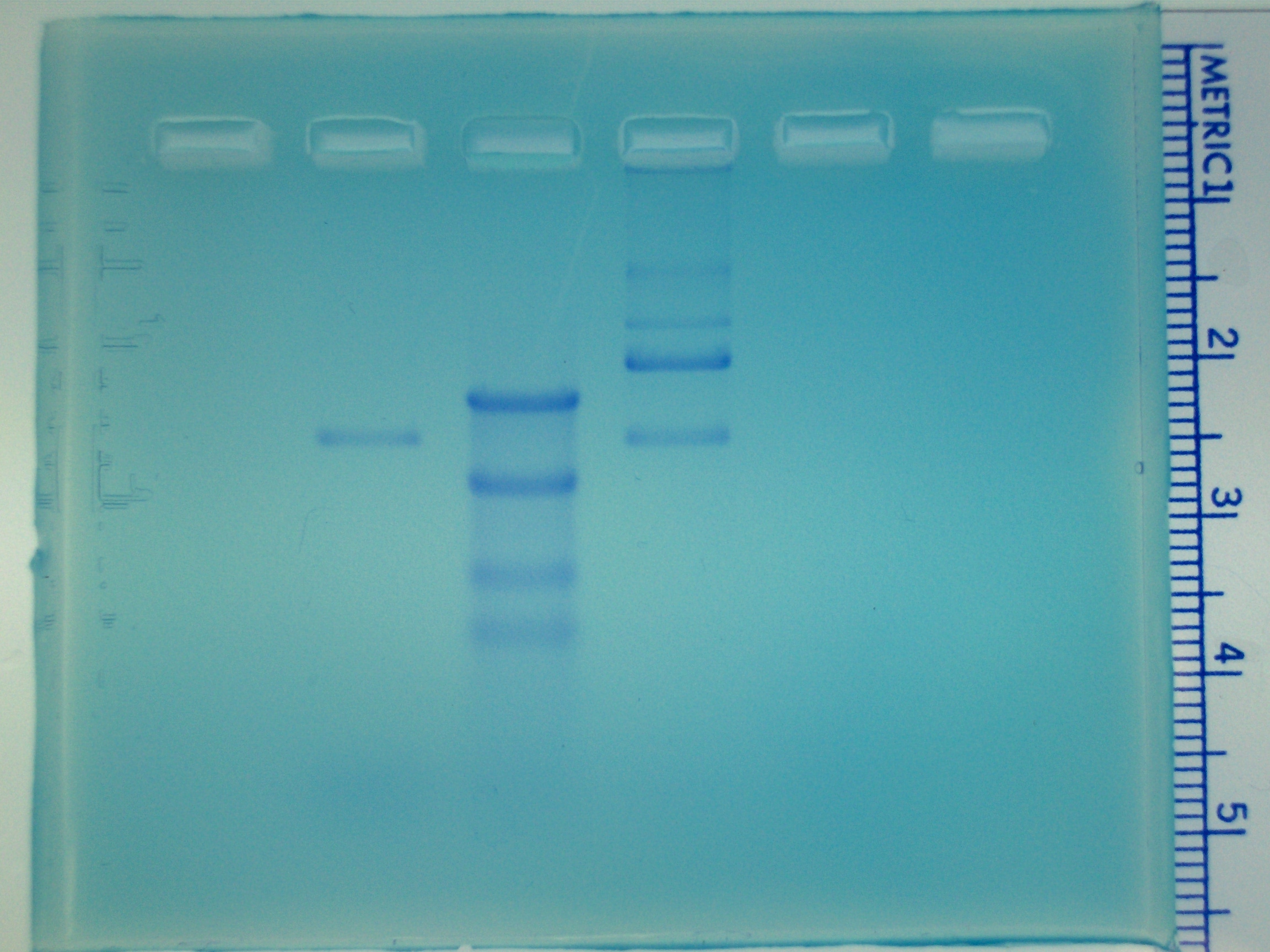 |
|
Agarose gel showing DNA bands stained with
methylene blue. Sample "A" was loaded in lane 2,
"DNA markers" were loaded in lane 3, sample "B" was loaded in lane 4, and
sample "C" was loaded in lane 5.
|
|
Your Turn |
| What is the main difference between
protein electrophoresis and DNA electrophoresis?
|
Check your answer. |
| How is the separation of DNA molecules by
agarose gel electrophoresis similar to the separation of proteins by SDS-PAGE electrophoresis?
|
Check your answer. |
| Which electrode do DNA molecules move
towards during electrophoresis? |
Check your answer. |
| Which factor is the main determinant of
how fast DNA molecules move through the gel during electrophoresis?
|
Check your answer. |
| When measuring the size of DNA molecules,
what units of measurement are generally used?
|
Check your answer. |
| How can agarose gel electrophoresis be
used to estimate the size of DNA molecules?
|
Check your answer. |
Close this browser window to return
to Blackboard and complete the practice quiz and assessment quiz.

